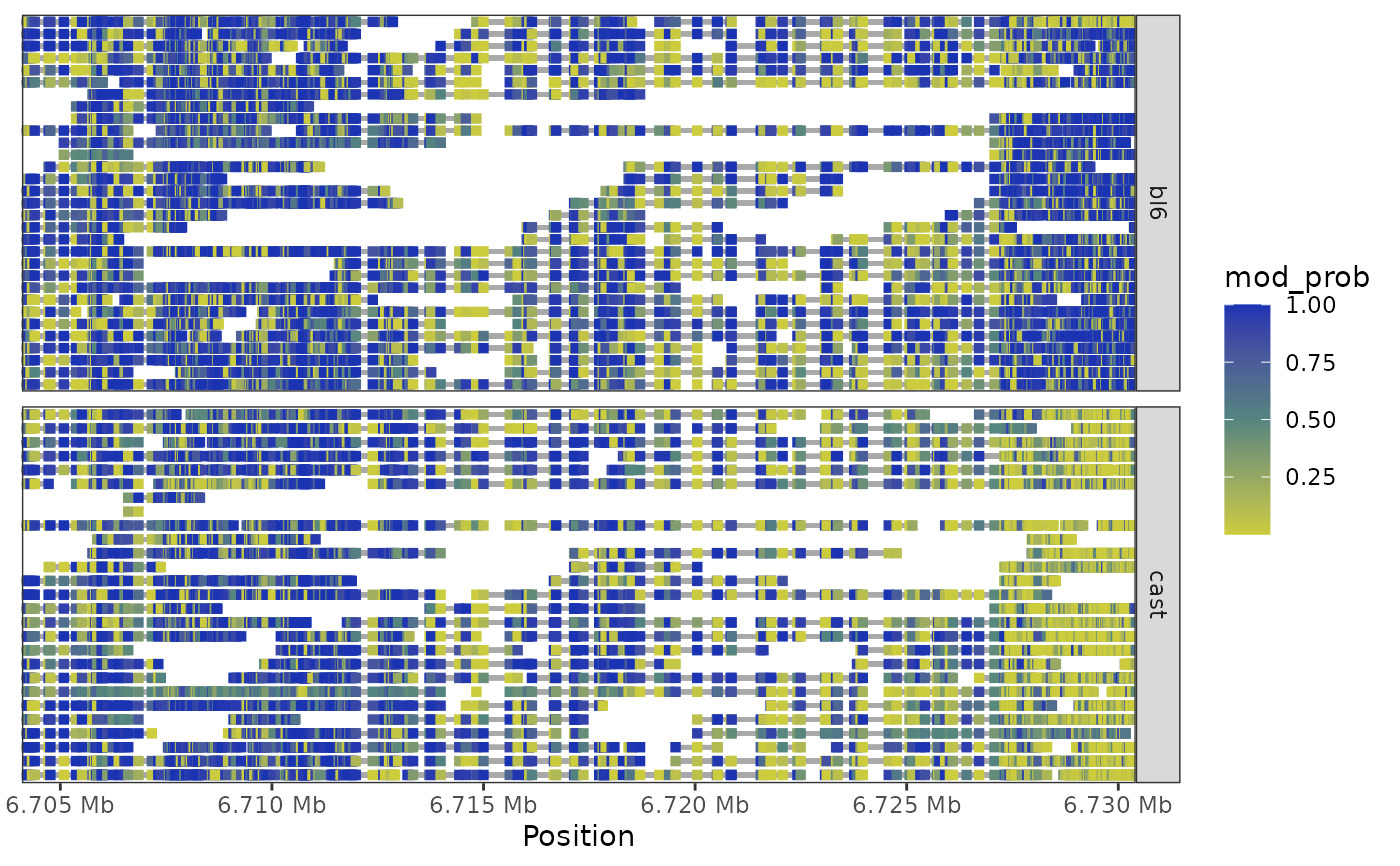
Plot GRanges heatmap
Usage
plot_grange_heatmap(
x,
grange,
pos_style = c("to_scale", "compact"),
window_prop = 0,
subsample = 50
)Arguments
- x
the NanoMethResult object.
- grange
the GRanges object with one entry.
- pos_style
the style for plotting the base positions along the x-axis. Defaults to "to_scale", plotting (potentially) overlapping squares along the genomic position to scale. The "compact" options plots only the positions with measured modification.
- window_prop
the size of flanking region to plot. Can be a vector of two values for left and right window size. Values indicate proportion of region length.
- subsample
the number of read of packed read rows to subsample to.
Examples
nmr <- load_example_nanomethresult()
#> Successfully matched 6 samples between data and annotation.
plot_grange_heatmap(nmr, GenomicRanges::GRanges("chr7:6703892-6730431"))