Chapter 1 Session 1
In this first session we will learn about:
- R and RStudio
- Data types and data structures
- Vectorised operations
- Data input and output
1.1 R and RStudio
R is a free and open source statistical programming language, great for performing data analysis. RStudio is a free and open source R integrated development environment (IDE) which makes it easier for you to write code. It does this by providing you with auto-completion (of variable names, function names, file paths etc.), helping with formatting and keeping track of your variables.
You can think of R as the engine in a car and RStudio as the body & controls. R is doing all the calculations/computations but RStudio makes it easier for you to use R.
When you first open RStudio, there will be three panels - see Figure 1.1 (don’t worry if your RStudio does not have the same colours/appearance - different versions of RStudio look slightly different, and the colour here is not the default one.)
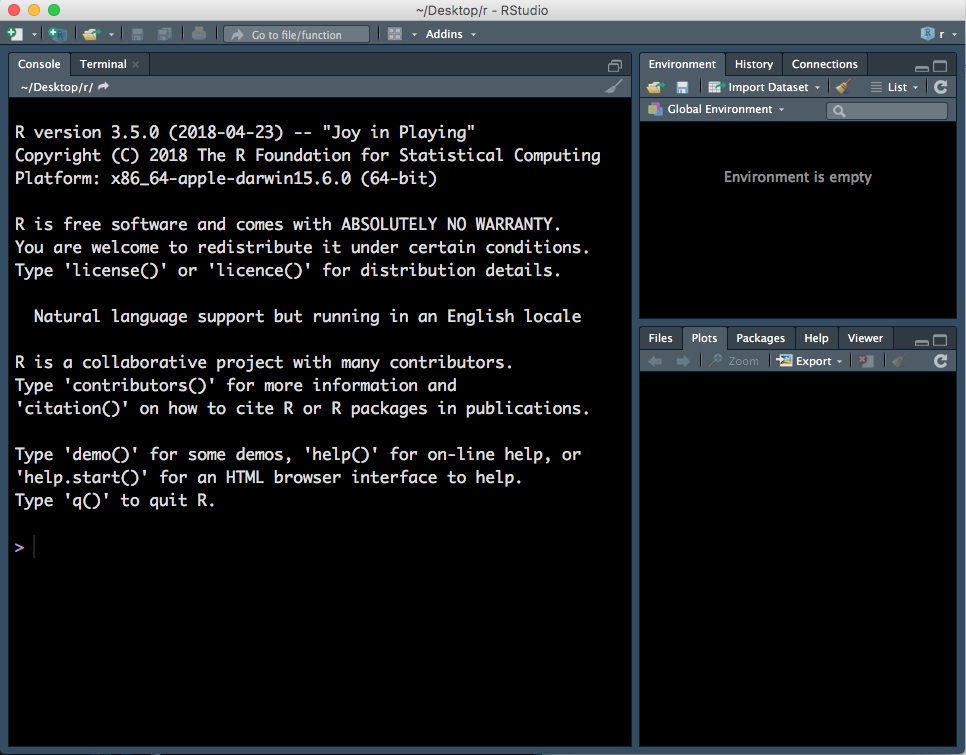
Figure 1.1: Three panels in RStudio.
- Left panel: this panel features two tabs, ‘Console’, which is where you can type in commands for R to run and ‘Terminal’, which we won’t worry about in this course.
- Top right panel:
- Environment - in this tab you can see all variables you have created.
- History - R keeps track of all commands you have run and you can review them in this tab.
- Connections - this tab helps you connect to data sources but we will not be using it in this course.
- Bottom right:
- Files - you can explore your file directory here and we will use it to set our working directory later.
- Plots - plots that you create will either appear here or be saved to a file.
- Help - help files for R functions can be viewed in this tab. Help files tell you about what a function does and how to use it.
- Packages - basic R includes many useful functions. You can add even more functions by downloading packages. A package is a collection of functions, generally with a certain data analysis theme. For example, the package ‘limma’, which we will use later, includes functions for analysing RNA-sequencing data.
- Viewer - this tab lets you view local web content but we won’t be using it in this course.
1.2 R scripts
To open an R script, go to File > New File > R Script.
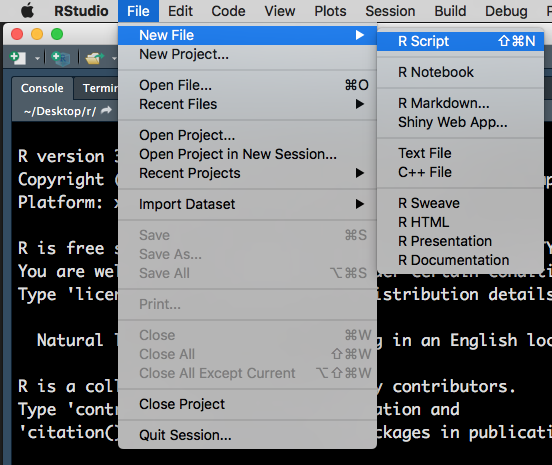
Figure 1.2: Opening a new R Script.
This will open a fourth panel on the top left.
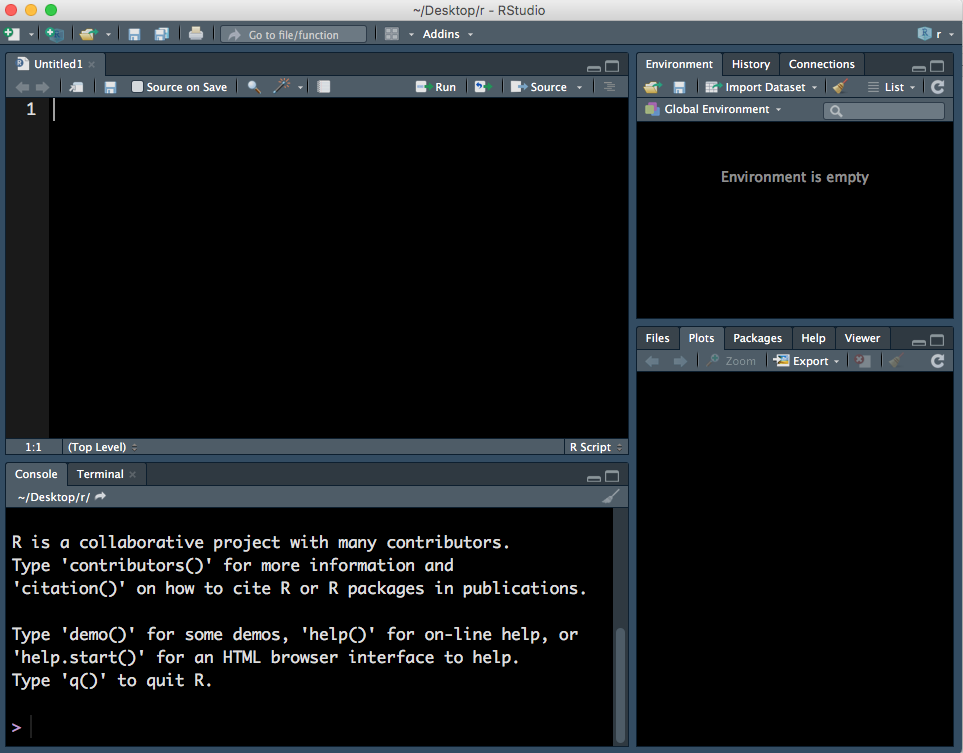
Figure 1.3: Four panels, including a R Script.
An R script is a text document where can type and run commands. You can also run commands in the console but the code run in the console is not saved. Note that to run a command in the Console press Enter key but to run a command in a R Script you must press Cmd/Ctrl + Enter keys.
Lastly, you add ‘comments’ in your R Script. Comments are notes regarding the code that are not interpreted by R, they begin with #:
## [1] 21.3 Working directory
Every file on your computer is located in a specific location. This location can be referred to by a path. In Mac, paths look something like this: /Users/Lucy/Documents/. In Windows, paths look something like this: C:\Users\Lucy\Documents\.
When you open an R session, it launches from a specific location. You can find out where this is using the command getwd(). This location is called the ‘working directory’. By default, R will look in this directory when reading in data and write out files/plots to this directory. It is often useful to have your data and R Scripts in the same directory and set this as your working directory.
You can set your working directory to be anywhere you like and we will now do this:
- Make a folder for this course, somewhere sensible on your computer that you will be able to easily find.
- Go back to your RStudio window, go to the bottom right panel, click on the ‘Files’ tab and then click on the three dots on the top right hand corner (Figure 1.4).
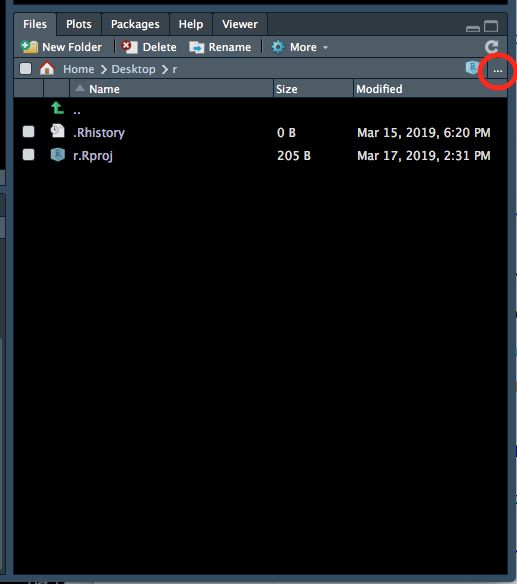
Figure 1.4: Setting the working directory - 1.
- This will open up a new window (Figure 1.5) which lets you explore the files and folders on your computer. Find the new folder you created, click on it then click ‘Open’.
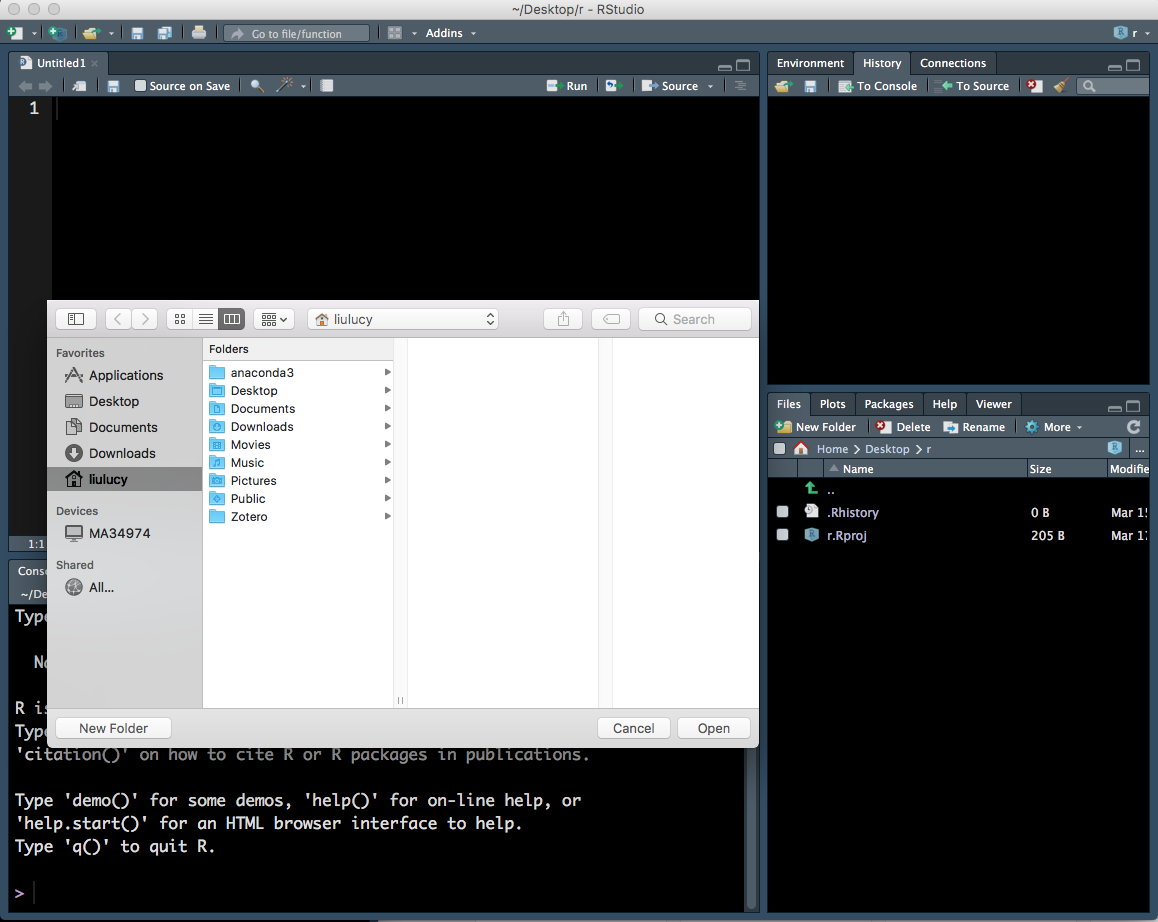
Figure 1.5: Setting the working directory - 2.
- The files tab will now show the contents of your new folder (which should be empty). At the top of the files tab, click on More > Set As Working Directory (Figure 1.6).
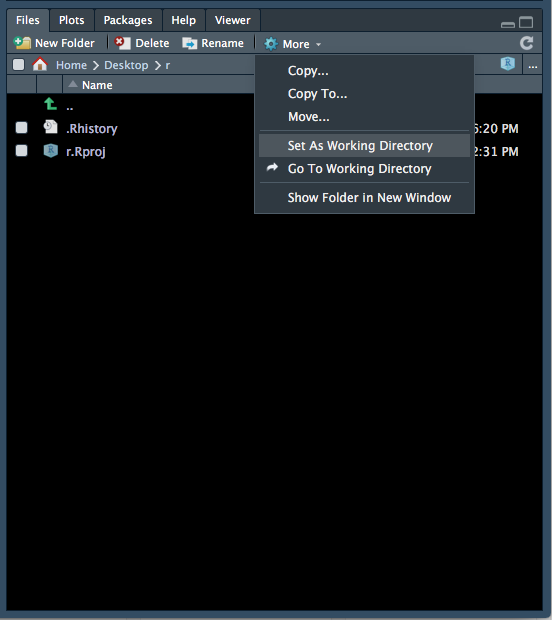
Figure 1.6: Setting the working directory - 3.
Please set your working directory to be this folder at the start of EVERY session.
1.4 Maths
R performs maths and follows standard order of operations. In order of highest to lowest precedence, here is how mathematical operations are denoted in R:
()- parentheses^or**- exponents/- divide*- multiply+- add-- subtract%%- remainder (modulus)
## [1] 3## [1] 13## [1] 16## [1] 11.5 Comparisons
R can perform comparisons:
==equal to.!=not equal to.>greater than.>=greater or equal to.<less than.<=less than or equal to.
These operations return a TRUE or a FALSE value. This can be used to quickly summarise data or perform indexing as we will see later.
## [1] FALSE## [1] TRUEYou can also compare words. R will use dictionary order to determine which word is ‘greater’.
## [1] FALSE1.6 Functions
Functions are expression in R that take inputs and produce outputs. Functions may take multiple inputs, also called ‘arguments’ to the function, each argument must be separated by comma. Arguments have a set order in which they can be given, or they can be referred to specifically by their name.
## [1] 3.141593## [1] 3.142Many arguments have default values so you don’t need to specify every argument for every function. You can check the arguments of a function by using args().
## function (n, mean = 0, sd = 1)
## NULLSometimes it’s obvious what the arguments are, but if more details are needed then you can access the help pages. This can be one in one of two ways, by searching for the function in the help panel.
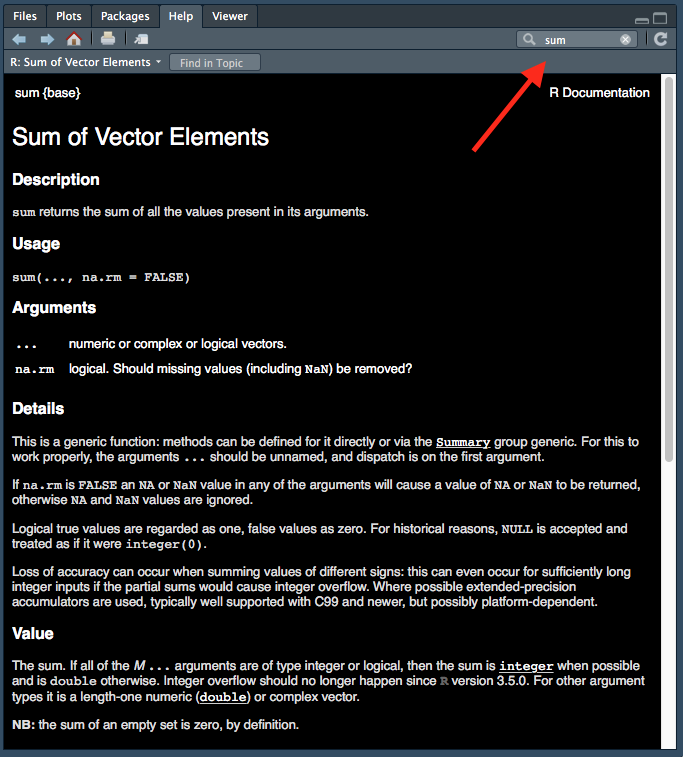
Figure 1.7: Accessing help files via ‘help’ tab.
Or using the command ? followed by the name of the function:
Help files are very useful but can be difficult to interpret at first due to the technical language used. It is often helpful to scroll down to the bottom to see examples of how the function is used.
1.7 Variables
A variable in R is an identifier to keep track of values. We assign values to variables so that we can refer to the variable later.
For example, I can do some maths:
## [1] 296R outputs simply the result. To use this value later, I would need to assign the output to a variable.
You can this with <- (shortcut = alt + -) in R. (You can also use =, however, stylistically <- is preferred.)
Here R first performs the calculation on the right of <- and then saves the result as a variable called my_num.
## [1] 296Note that variable names are case sensitive.
## Error in eval(expr, envir, enclos): object 'VAR' not foundYou can also ‘overwrite’ variables by assigning to them again:
## [1] 12Because the right hand side is evaluated first, you can also assign to a variable a calculation that involves itself.
## [1] 17R has rules on valid variable names. Variable names can only contain letters, numbers, the dot or underline characters. They can only start with a letter or the dot followed by a letter.
If we try to create a variable that starts with a number, R will return an error:
## Error: <text>:1:2: unexpected symbol
## 1: 2myvar
## ^Challenge 1.1
- Guess the output the result of the following expression then evaluate them to check.
## [1] 8## [1] 36## [1] TRUE## [1] TRUE## [1] 1- Which of the following are valid variable names? Those that are not, why are they not valid?
- var
- var2
- 2var
- var-2
- var.2
- What is the value of
zonce this code has run?
1.8 Data types
Every variable in R has a ‘type’. The type tells R what kind of data it is and consequently what it can and can’t do with the data. For example, it makes sense to perform mathematical functions on numbers but not on words.
There are three basic types of data in R:
logical- either TRUE or FALSEnumeric- numberscharacter- text surrounded by quotation marks
These are called ‘atomic’ data types as they are the most basic types of data from which other data types usually derive.
You can find the type of a variable using the class() function.
## [1] 1## [1] "1"## [1] "logical"## [1] "numeric"## [1] "character"## [1] "character"## [1] "character"## [1] 2## Error in "1" + "1": non-numeric argument to binary operator## [1] FALSE## Error in !"TRUE": invalid argument type1.8.1 Factors
Factors are a special type for encoding categorical variables. They are composed of the ‘values’ and the ‘levels’, externally they look like characters variables, but internally they are a series of integers denoting the group number and a separate series of characters denoting the identity of the groups.
## [1] sunny rainy rainy cloudy sunny
## Levels: cloudy rainy sunny## [1] "cloudy" "rainy" "sunny"## [1] 3 2 2 1 3Factors are generally a more efficient way of storing categorical variables, it stores only the level value as an integer and a single instance of each unique level. It also makes it possible to quickly re-encode the levels.
## [1] Sunny Rainy Rainy Cloudy Sunny
## Levels: Cloudy Rainy SunnyWe can set the order of the levels if we would like using the levels argument.
## [1] low medium medium high low
## Levels: low high mediumSetting these levels can change the behaviour of the variable in models or plots.
1.9 Data structures
Data structures are ways to organise many values without having to assign individual names to each value. They help us keep related data together in a single variable and offer powerful ways for us to interact with that data.
Three commonly used data structures are summarised below:
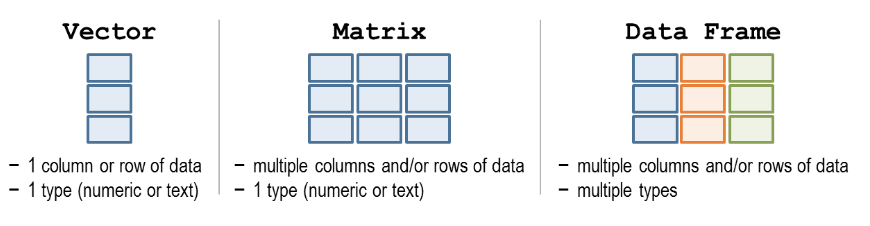
Figure 1.8: Data structures. Source: http://libguides.wellesley.edu.
Another useful data structure is a ‘list’, which we will talk about in Session 3.
1.9.1 Vector
A vector is a collection of values of the same type. Values in a vector are laid out linearly, one after another.
You can create vectors with the c() function (‘c’ for ‘combine’):
my_vect1 <- c(1, 2, 3) # a vector of numbers
my_vect2 <- c("a", "vector", "of", "characters") # a vector of charactersOne useful shorthand for creating a sequence of integers is a:b which creates a vector from a to b.
## [1] 1 2 3 4 5 6 7 8 9 10## [1] 3 4 5 6 7For very long vectors, it is useful to use the head and tail functions to inspect just a few values. These print the first and last 6 elements of a vector.
## [1] 1 2 3 4 5 6## [1] 9995 9996 9997 9998 9999 100001.9.2 Matrix
A matrix is the two-dimensional extension of the vector, it stores a collection of values of the same type but laid out in a grid with rows and columns. An example of this is a gene count matrices where each genes are represented by the rows, samples are represented by the columns and each cell represents a count for a particular gene in a particular column. The rows and columns can also be labelled with names, but these names are usually considered metadata rather than being a part of the matrix.
gene_counts <- matrix(c(1, 2, 3, 4), nrow = 2, ncol = 2)
rownames(gene_counts) <- c("gene1", "gene2")
colnames(gene_counts) <- c("sample1", "sample2")
class(gene_counts)## [1] "matrix" "array"## sample1 sample2
## gene1 1 3
## gene2 2 41.9.3 Data Frame
Data frames are similar to matrices in that they store data in rows and columns. The difference is that the data within each column can be of different types. This is the representation used for general tables that record different attributes of individual entries along the rows.
A classic example of a data frame is the iris dataset.
## [1] "data.frame"## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## 11 5.4 3.7 1.5 0.2 setosa
## 12 4.8 3.4 1.6 0.2 setosa
## 13 4.8 3.0 1.4 0.1 setosa
## 14 4.3 3.0 1.1 0.1 setosa
## 15 5.8 4.0 1.2 0.2 setosa
## 16 5.7 4.4 1.5 0.4 setosa
## 17 5.4 3.9 1.3 0.4 setosa
## 18 5.1 3.5 1.4 0.3 setosa
## 19 5.7 3.8 1.7 0.3 setosa
## 20 5.1 3.8 1.5 0.3 setosa
## 21 5.4 3.4 1.7 0.2 setosa
## 22 5.1 3.7 1.5 0.4 setosa
## 23 4.6 3.6 1.0 0.2 setosa
## 24 5.1 3.3 1.7 0.5 setosa
## 25 4.8 3.4 1.9 0.2 setosa
## 26 5.0 3.0 1.6 0.2 setosa
## 27 5.0 3.4 1.6 0.4 setosa
## 28 5.2 3.5 1.5 0.2 setosa
## 29 5.2 3.4 1.4 0.2 setosa
## 30 4.7 3.2 1.6 0.2 setosa
## 31 4.8 3.1 1.6 0.2 setosa
## 32 5.4 3.4 1.5 0.4 setosa
## 33 5.2 4.1 1.5 0.1 setosa
## 34 5.5 4.2 1.4 0.2 setosa
## 35 4.9 3.1 1.5 0.2 setosa
## 36 5.0 3.2 1.2 0.2 setosa
## 37 5.5 3.5 1.3 0.2 setosa
## 38 4.9 3.6 1.4 0.1 setosa
## 39 4.4 3.0 1.3 0.2 setosa
## 40 5.1 3.4 1.5 0.2 setosa
## 41 5.0 3.5 1.3 0.3 setosa
## 42 4.5 2.3 1.3 0.3 setosa
## 43 4.4 3.2 1.3 0.2 setosa
## 44 5.0 3.5 1.6 0.6 setosa
## 45 5.1 3.8 1.9 0.4 setosa
## 46 4.8 3.0 1.4 0.3 setosa
## 47 5.1 3.8 1.6 0.2 setosa
## 48 4.6 3.2 1.4 0.2 setosa
## 49 5.3 3.7 1.5 0.2 setosa
## 50 5.0 3.3 1.4 0.2 setosa
## 51 7.0 3.2 4.7 1.4 versicolor
## 52 6.4 3.2 4.5 1.5 versicolor
## 53 6.9 3.1 4.9 1.5 versicolor
## 54 5.5 2.3 4.0 1.3 versicolor
## 55 6.5 2.8 4.6 1.5 versicolor
## 56 5.7 2.8 4.5 1.3 versicolor
## 57 6.3 3.3 4.7 1.6 versicolor
## 58 4.9 2.4 3.3 1.0 versicolor
## 59 6.6 2.9 4.6 1.3 versicolor
## 60 5.2 2.7 3.9 1.4 versicolor
## 61 5.0 2.0 3.5 1.0 versicolor
## 62 5.9 3.0 4.2 1.5 versicolor
## 63 6.0 2.2 4.0 1.0 versicolor
## 64 6.1 2.9 4.7 1.4 versicolor
## 65 5.6 2.9 3.6 1.3 versicolor
## 66 6.7 3.1 4.4 1.4 versicolor
## 67 5.6 3.0 4.5 1.5 versicolor
## 68 5.8 2.7 4.1 1.0 versicolor
## 69 6.2 2.2 4.5 1.5 versicolor
## 70 5.6 2.5 3.9 1.1 versicolor
## 71 5.9 3.2 4.8 1.8 versicolor
## 72 6.1 2.8 4.0 1.3 versicolor
## 73 6.3 2.5 4.9 1.5 versicolor
## 74 6.1 2.8 4.7 1.2 versicolor
## 75 6.4 2.9 4.3 1.3 versicolor
## 76 6.6 3.0 4.4 1.4 versicolor
## 77 6.8 2.8 4.8 1.4 versicolor
## 78 6.7 3.0 5.0 1.7 versicolor
## 79 6.0 2.9 4.5 1.5 versicolor
## 80 5.7 2.6 3.5 1.0 versicolor
## 81 5.5 2.4 3.8 1.1 versicolor
## 82 5.5 2.4 3.7 1.0 versicolor
## 83 5.8 2.7 3.9 1.2 versicolor
## 84 6.0 2.7 5.1 1.6 versicolor
## 85 5.4 3.0 4.5 1.5 versicolor
## 86 6.0 3.4 4.5 1.6 versicolor
## 87 6.7 3.1 4.7 1.5 versicolor
## 88 6.3 2.3 4.4 1.3 versicolor
## 89 5.6 3.0 4.1 1.3 versicolor
## 90 5.5 2.5 4.0 1.3 versicolor
## 91 5.5 2.6 4.4 1.2 versicolor
## 92 6.1 3.0 4.6 1.4 versicolor
## 93 5.8 2.6 4.0 1.2 versicolor
## 94 5.0 2.3 3.3 1.0 versicolor
## 95 5.6 2.7 4.2 1.3 versicolor
## 96 5.7 3.0 4.2 1.2 versicolor
## 97 5.7 2.9 4.2 1.3 versicolor
## 98 6.2 2.9 4.3 1.3 versicolor
## 99 5.1 2.5 3.0 1.1 versicolor
## 100 5.7 2.8 4.1 1.3 versicolor
## 101 6.3 3.3 6.0 2.5 virginica
## 102 5.8 2.7 5.1 1.9 virginica
## 103 7.1 3.0 5.9 2.1 virginica
## 104 6.3 2.9 5.6 1.8 virginica
## 105 6.5 3.0 5.8 2.2 virginica
## 106 7.6 3.0 6.6 2.1 virginica
## 107 4.9 2.5 4.5 1.7 virginica
## 108 7.3 2.9 6.3 1.8 virginica
## 109 6.7 2.5 5.8 1.8 virginica
## 110 7.2 3.6 6.1 2.5 virginica
## 111 6.5 3.2 5.1 2.0 virginica
## 112 6.4 2.7 5.3 1.9 virginica
## 113 6.8 3.0 5.5 2.1 virginica
## 114 5.7 2.5 5.0 2.0 virginica
## 115 5.8 2.8 5.1 2.4 virginica
## 116 6.4 3.2 5.3 2.3 virginica
## 117 6.5 3.0 5.5 1.8 virginica
## 118 7.7 3.8 6.7 2.2 virginica
## 119 7.7 2.6 6.9 2.3 virginica
## 120 6.0 2.2 5.0 1.5 virginica
## 121 6.9 3.2 5.7 2.3 virginica
## 122 5.6 2.8 4.9 2.0 virginica
## 123 7.7 2.8 6.7 2.0 virginica
## 124 6.3 2.7 4.9 1.8 virginica
## 125 6.7 3.3 5.7 2.1 virginica
## 126 7.2 3.2 6.0 1.8 virginica
## 127 6.2 2.8 4.8 1.8 virginica
## 128 6.1 3.0 4.9 1.8 virginica
## 129 6.4 2.8 5.6 2.1 virginica
## 130 7.2 3.0 5.8 1.6 virginica
## 131 7.4 2.8 6.1 1.9 virginica
## 132 7.9 3.8 6.4 2.0 virginica
## 133 6.4 2.8 5.6 2.2 virginica
## 134 6.3 2.8 5.1 1.5 virginica
## 135 6.1 2.6 5.6 1.4 virginica
## 136 7.7 3.0 6.1 2.3 virginica
## 137 6.3 3.4 5.6 2.4 virginica
## 138 6.4 3.1 5.5 1.8 virginica
## 139 6.0 3.0 4.8 1.8 virginica
## 140 6.9 3.1 5.4 2.1 virginica
## 141 6.7 3.1 5.6 2.4 virginica
## 142 6.9 3.1 5.1 2.3 virginica
## 143 5.8 2.7 5.1 1.9 virginica
## 144 6.8 3.2 5.9 2.3 virginica
## 145 6.7 3.3 5.7 2.5 virginica
## 146 6.7 3.0 5.2 2.3 virginica
## 147 6.3 2.5 5.0 1.9 virginica
## 148 6.5 3.0 5.2 2.0 virginica
## 149 6.2 3.4 5.4 2.3 virginica
## 150 5.9 3.0 5.1 1.8 virginicaThe head() and tail() functions also work here to print the first and last 6 rows of a data frame for a quick check of the data.
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 145 6.7 3.3 5.7 2.5 virginica
## 146 6.7 3.0 5.2 2.3 virginica
## 147 6.3 2.5 5.0 1.9 virginica
## 148 6.5 3.0 5.2 2.0 virginica
## 149 6.2 3.4 5.4 2.3 virginica
## 150 5.9 3.0 5.1 1.8 virginicaWe can construct a data frame of our own as well using data.frame()
my_df <- data.frame(
mouse_id = c("mouse1", "mouse2", "mouse3"),
age = c(48, 48, 52),
weight = c(39.7, 42.2, 46.3)
)
class(my_df)## [1] "data.frame"## mouse_id age weight
## 1 mouse1 48 39.7
## 2 mouse2 48 42.2
## 3 mouse3 52 46.3The general form for constructing a data frame looks like this
data.frame(col_name1 = values1, col_names2 = values2, ...)Where each value is a vector of the same length.
You can access a column in a data frame with the shortcut $. Notice that the names of all columns of the data frame appears after typing in my_df$:
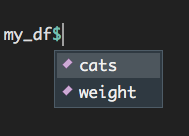
Figure 1.9: Accessing columns in a dataframe using the ‘$’ symbol.
1.10 Reading in data
Let’s read in some data and start exploring it.
You should have received the data files via email before the course. Please download these files and make sure they are located in your working directory. Recall, we set our working directory above. You can find the location of your working directory with the function getwd().
If you are following along outside of the WEHI course, you can download the data files from Github - instructions for downloading data from GitHub can be found in the Preface.
The file we want to read in is named ‘Ses1_genes.tsv’.
All the data files are in a directory called ‘data’ - thus the path to the file (relative to my working directory) is ‘data/Ses1_genes.tsv’. Depending on where you have put your data (in your working directory or in another file in your working directory), the path to your file ‘Ses1_genes.tsv’ may be different.
## SYMBOL GeneLength Count
## 1 Gm10568 1634 0
## 2 Gm19860 799 4
## 3 Gm19938 3259 0
## 4 Lypla1 2433 768
## 5 Rp1 9747 0
## 6 Sox17 3130 1
## 7 Tcea1 2847 810
## 8 Mrpl15 4203 431
## 9 Xkr4 3634 1
## 10 Rgs20 2241 452Note that read.delim() also lets you specify character that separates columns of the data. The most common types are comma-separated values (csv) and tab-separated values (tsv).
Example of a csv file:
Name, Age
Andy, 10
Bob, 8Example of a tsv file:
Name Age
Andy 10
Bob 8By default read.delim() uses the tab separator ". We can see what happens if we use the wrong separator in our read.delim call.
## SYMBOL.GeneLength.Count
## 1 Gm10568\t1634\t0
## 2 Gm19860\t799\t4
## 3 Gm19938\t3259\t0
## 4 Lypla1\t2433\t768
## 5 Rp1\t9747\t0
## 6 Sox17\t3130\t1
## 7 Tcea1\t2847\t810
## 8 Mrpl15\t4203\t431
## 9 Xkr4\t3634\t1
## 10 Rgs20\t2241\t452If you just read in the data, R simply prints the values out in the console. Let’s assign our data frame to a variable called genes:
Notice how genes now appears in our ‘Environment’ tab:
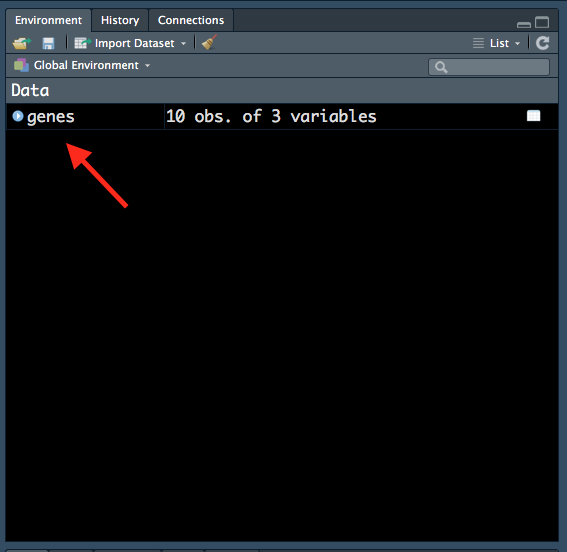
Figure 1.10: Variable in ‘Environment’ tab.
All variables that we create will be shown in this tab, so it is a useful way to keep track of variables that we have created. Notice how R also tells us that there are 10 observations (rows) and 3 variables (columns) in the genes data frame.
One quirk that older versions of R has is that columns of character strings are automatically converted to factors. Factors look like characters when printed out, but can cause errors if you actually treat them as characters. It’s generally wise to turn off this automatic conversion. If your R is newer than version 4.0.0 then stringsAsFactors is off by default.
genes <- read.delim("data/Ses1_genes.tsv", stringsAsFactors = TRUE)
str(genes) # the SYMBOL column is of type factor## 'data.frame': 10 obs. of 3 variables:
## $ SYMBOL : Factor w/ 10 levels "Gm10568","Gm19860",..: 1 2 3 4 7 8 9 5 10 6
## $ GeneLength: int 1634 799 3259 2433 9747 3130 2847 4203 3634 2241
## $ Count : int 0 4 0 768 0 1 810 431 1 452genes <- read.delim("data/Ses1_genes.tsv", stringsAsFactors = FALSE)
str(genes) # the SYMBOL column is now of type character## 'data.frame': 10 obs. of 3 variables:
## $ SYMBOL : chr "Gm10568" "Gm19860" "Gm19938" "Lypla1" ...
## $ GeneLength: int 1634 799 3259 2433 9747 3130 2847 4203 3634 2241
## $ Count : int 0 4 0 768 0 1 810 431 1 4521.11 Vectorised Operations
In contrast to some other programming languages where operating on vectors requires a loop structure, operations in R are generally vectorised, meaning they will be automatically applied to each element of the vector.
## [1] 0 4 0 768 0 1 810 431 1 452## [1] 10 14 10 778 10 11 820 441 11 462## [1] TRUE FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE FALSE## [1] TRUE TRUE TRUE FALSE TRUE TRUE FALSE FALSE TRUE FALSEThis can be useful for creating new columns based on existing data
When assigning in this way, an existing column may be overwritten. If the column assigned to does not exist, a new column is created.
Challenge 1.3
Create a new column called Prop_Count that contains each count value as a proportion of the total count value of all 10 genes in the data frame. E.g. if Count was 10 and total count of all 10 genes is 100, that row in Prop_Count should be 0.1.
Hint use the sum() function.
1.12 Writing out data
The last thing we will do this session is learn to write out data using the function write.table().
There are a few things we must tell write.table(), for it to be able to write out the data the way we want:
x- the name of this input is not very informative, but first you must tell the function what you want to write out. In our case we want to write out our dataframegenes.file- the name of the file that we want to write to.sep- how each value in our output file is separated. Common file formats are ‘csv’ and ‘tsv’ (discussed above). In R, a tab is represented by"\t".row.names- this is eitherTRUEorFALSE, and let’s you specify whether you want to write out row names. If your dataframe does not have row names, putFALSE.col.names- this is also eitherTRUEorFALSE, and let’s you specify whether you want to write out column names. If your dataframe has column names, putTRUE.
We can write out our genes data frame into a .tsv file using the command below: